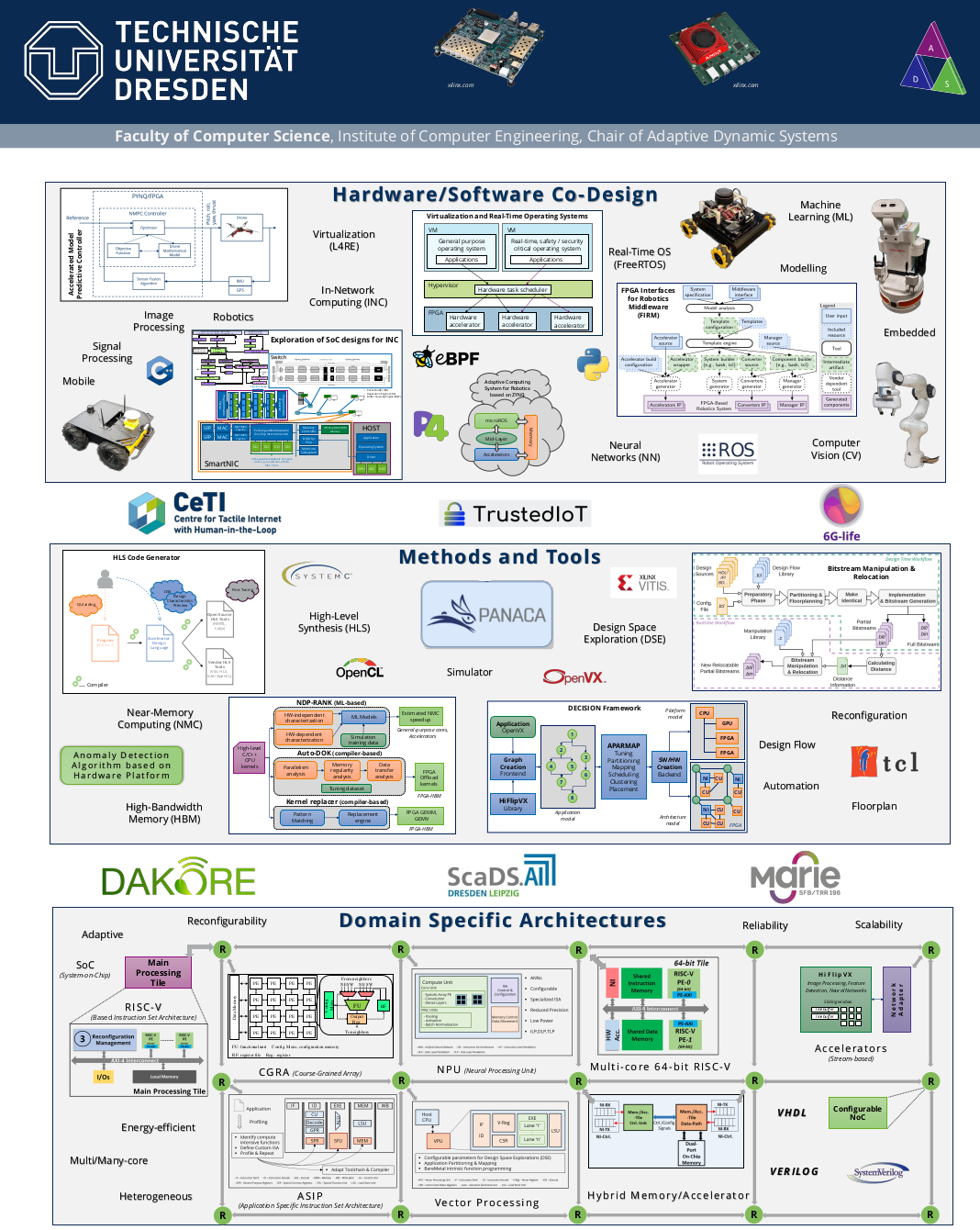
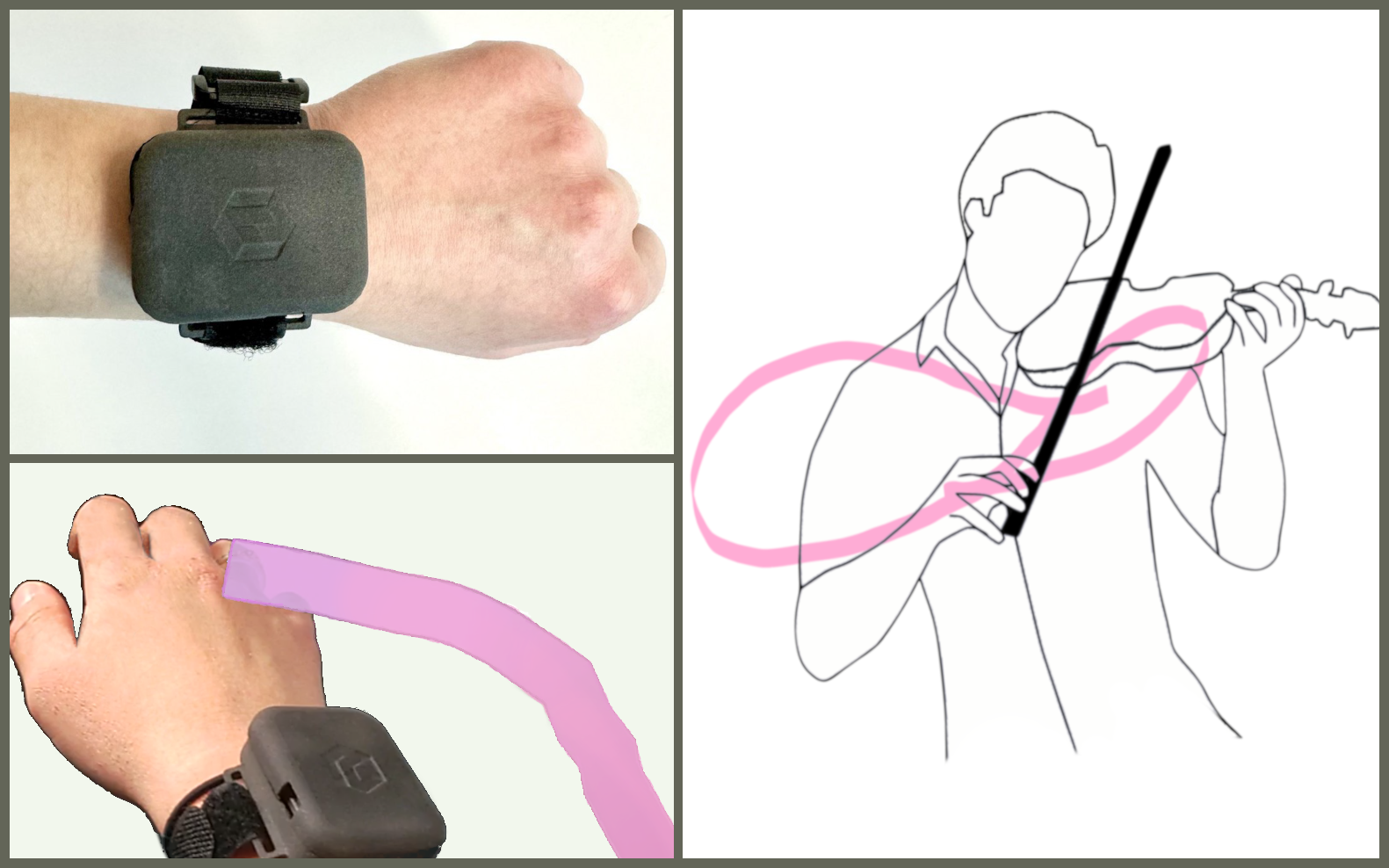
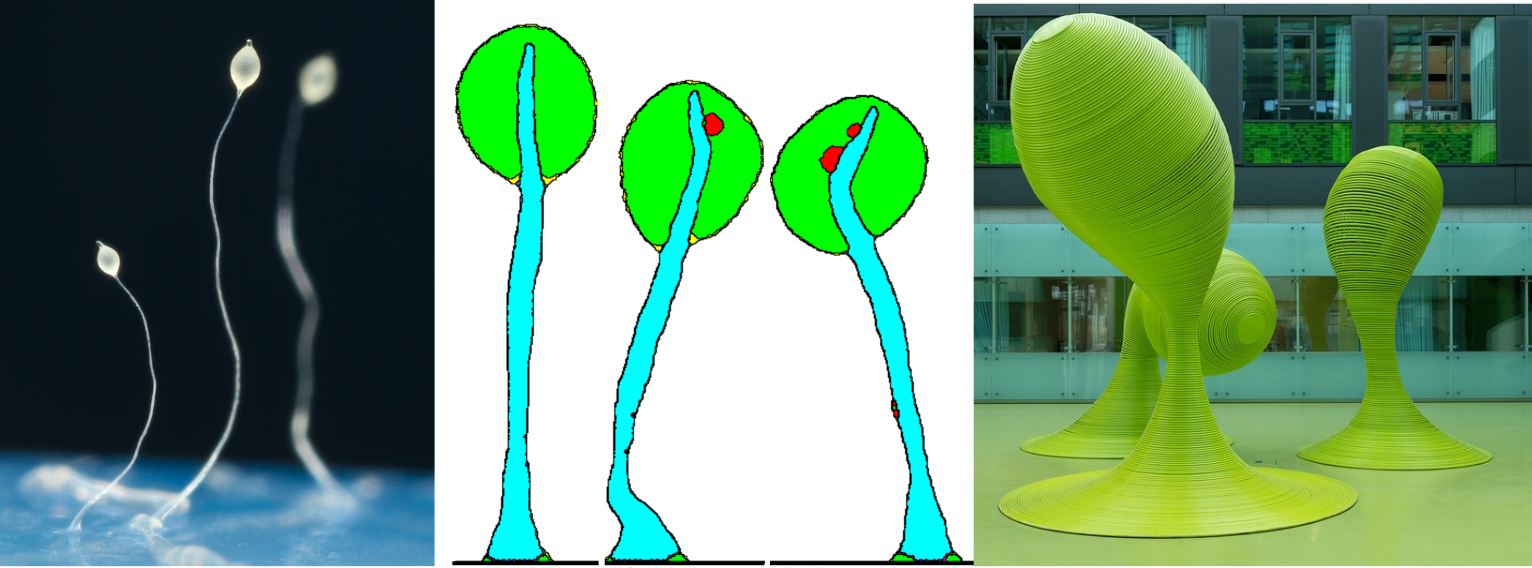
DIE PROJEKTSCHAU DER FAKULTÄT INFORMATIK AN DER TU DRESDEN
Einmal im Jahr präsentieren Studenten und Mitarbeiter ihre Ergebnisse aus Lehre und Forschung der Öffentlichkeit. Dich erwarten spannende Installationen, Workshops, Vorträge und Ausstellungen. Zudem bietet OUTPUT.DD die Gelegenheit, mit Unternehmen über aktuelle Forschungsfragen ins Gespräch zu kommen.
Die Veranstaltung ist offen für alle.
Wir freuen uns auf dich!